How I Built a Colorectal Cancer Prediction Platform through Deep Learning
Last Updated on August 26, 2020 by Editorial Team
Author(s): Adrian Serapio
Deep Learning

I believe that artificial intelligence can save the human race.
How ironic is it though that as any generic sci-fi portrays it, any advanced AI would suddenly conceive the thought that exterminating the human race is the solution for world peace.

A Skynet fantasy is kinda remote from happening… for now at least.
Kidding aside, instead of focusing on the 0.001% probability that artificial intelligence will lead to the destruction of mankind, it would be worthwhile to consider that AI could be very beneficial to society, especially in the healthcare sector.

The applications of AI in the field of healthcare are limitless, offering huge potential in the field of precision medicine, health analytics, medical informatics, etc.
In my case, I tried to investigate the potential of artificial intelligence in the field of oncology. I sought to leverage the power of deep learning to build neural networks trained on the cloud to more accurately and efficiently determine if a colorectal tissue contains tumors or not.
Why Cloudiopsy?
Colorectal cancer (CRC) claimed approximately 1.8 million lives in the year 2018 alone (WHO, 2018). In fact, in the Philippines, colorectal cancer is deemed as the number one gastrointestinal cancer (Afinidad-Bernardo, 2017). Fortunately, however, colorectal cancer is preventable and when identified at an early age, colorectal cancer could be cured.
As such, Cloudiopsy is designed as a colorectal cancer prediction platform that could be easily distributed in the country to aid in diagnosis by differentiating between normal mucosal tissue and epithelial adenocarcinoma (tumor) tissue. This is a cost-efficient way of making colorectal cancer prediction universally available in the Philippines, given that the diagnosis platform could be in the future, easily accessible online.
So, what is it?
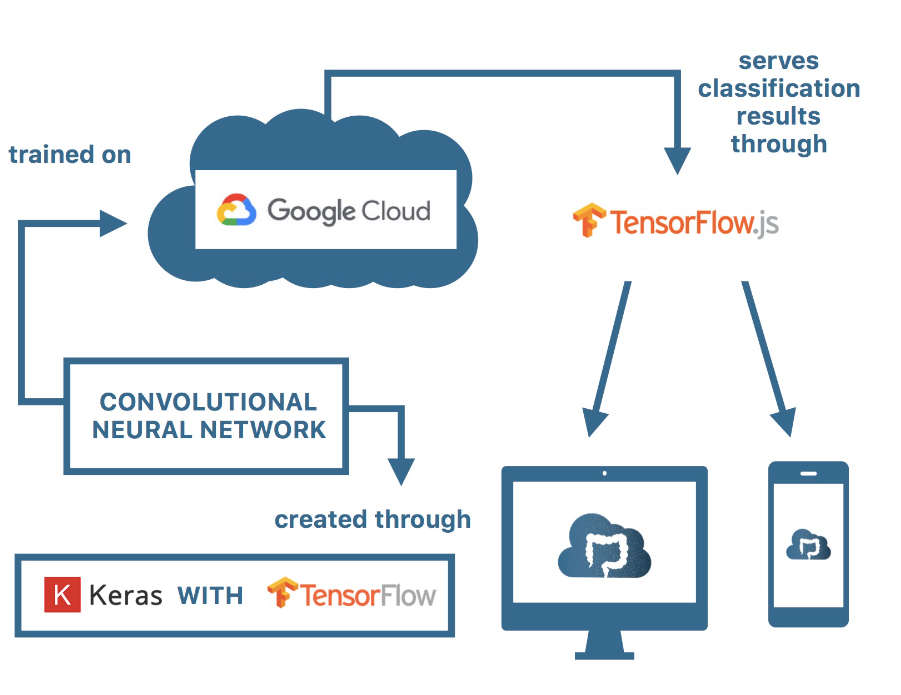
Put simply, the Cloudiopsy platform predicts image inputs through a neural network model through transfer learning, using Keras with a Tensorflow backend trained on Google Cloud Platform and serving predictions in the browser through Tensorflow.js.
Basic Pathological Intuition behind Colorectal Cancer
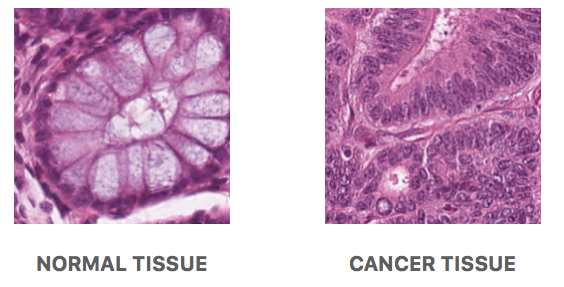
The picture on the left is an example of a normal mucosal tissue layer. It could be seen that goblet cells (mucus-secreting cells) are relatively the same size and populate the colonic crypt (an intestinal gland which is elliptical in shape for this cross-section). This indicates that the crypt is healthy, and thus, this picture was taken from normal mucosal tissue.
As opposed to the picture on the right, the goblet cells are no longer visible inside of the tissue which would imply it already is tumor tissue. This type of tumor is specifically known as epithelial adenocarcinoma. This could be an indicator of colorectal cancer if other region samples sent are also proven to be containing tumors.
Current methods of analyzing tissue samples given biopsies are to manually scan a sample and find for cancerous regions by looking at the tissue’s architecture. However, scanning through the entire region is very laborious and is prone to human error. As such, it would be ideal to automate this process and find a way to create a platform that will automatically determine if a patch is of normal tissue or cancer tissue and return predictions.
Neural Networks and Transfer Learning
Neural networks are a set of connections in the brain where electrical signals are fired through each other to generate thoughts, ideas, emotions, etc. On the other hand, when we learn, we re-arrange these connections between these neurons to learn new patterns and concepts.
Modeled after our brain, neural networks can transform information through mathematical operations to obtain a specific output. As such, neural networks have the capacity to translate complicated patterns such as image or speech into numerical values and mathematically transform these values through multiplying by weights, to classify them.
Weights are simply values in which the input is multiplied or transformed by. These weights are what is modified until a specific output (classification or prediction) is generated.
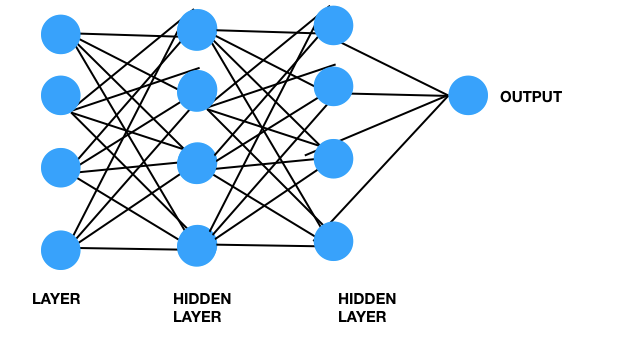
Moreover, in order to capture patterns and “learn” which one is a tumour and which is normal, certain layers in the neural network may be responsible for learning certain features regarding the image. Initial layers may be responsible for identifying features such as edges, and corners, and moving along the neural network, some layers go through higher levels of abstractions such as understanding of shapes (circles vs ellipses), and later on, possibly distance between these, etc.
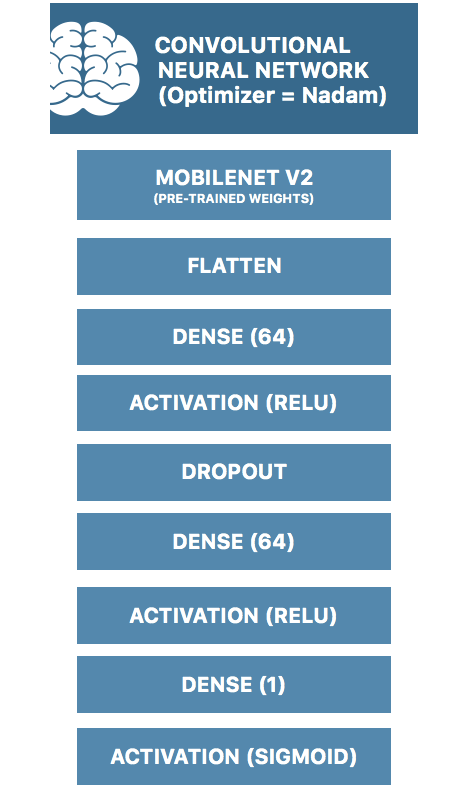
One method would be to train a neural network from scratch. However, in my project, instead of training from scratch, knowing that the lower layers may be responsible for edge detection and shape detection, I have used transfer learning wherein I have initialized it with pre-trained weights from pre-trained neural networks. Specifically, I have pre-trained on the Mobilenet V2 model.
The Dataset
The dataset utilized for this project was the 100,000 histological images of human colorectal cancer and healthy tissue prepared by Kather, Halama, and Marx. According to the researchers, the dataset is composed of 100,000 images of hematoxylin & eosin (H&E) stained histological images of human colorectal cancer (CRC) and normal tissue collated from samples from the NCT Biobank and the UMM pathology archive (Kather, Halama & Marx, 2016).
Building the Model
To build the neural network model, I have utilized the Google Cloud Platform to train on over 4534 images (2235 images of tumor tissue and 2299 images of normal tissue). I have 1330 images for validation (656 for normal and 674 for tumorous tissue) and 1653 images (810 of normal and 843 of tumor tissue) for a test dataset.
I have utilized Keras with Tensorflow backend to design the neural network with a Nadam optimizer, training for 50 epochs.
The specific architecture for my model is illustrated below:
Results
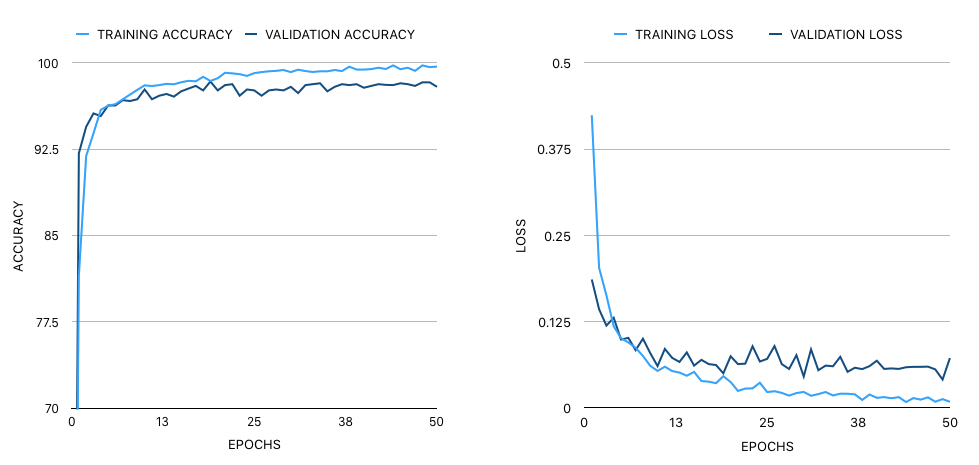
The loss function was based on binary cross-entropy. The model gained a final test accuracy of 98.79% and an F1 score of 0.987. The final size of the model is 75.5 MB.
Serving using Tensorflow.js
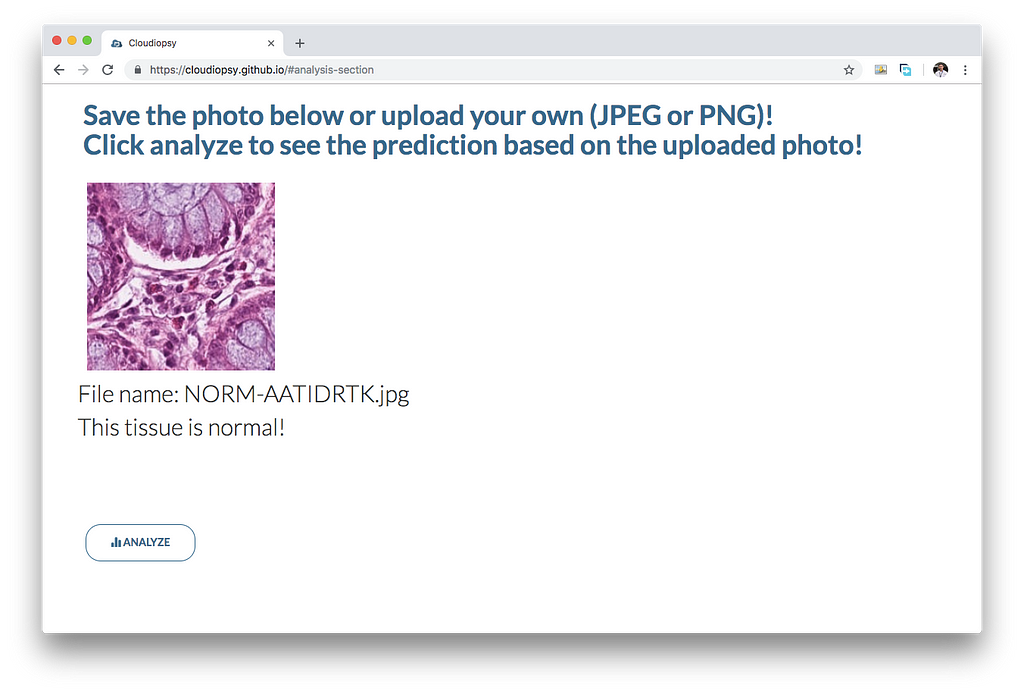
In order to serve it online, I have utilized the tfjs converter, which converts the model into a model.json file and shard files which will be used to load the model. I have written a simple script to load the model.json and serve predictions through cloudiopsy.github.io.
Conclusion
This is still a work of progress as a higher accuracy is more favorable to create a tool that will be very useful for pathologists and medical professionals. Moreover, testing on the clinical setting would reveal possible refinements in the software. However, one day, Cloudiopsy may hopefully be used to help diagnose colorectal cancer through a faster analysis of colorectal biopsies and aid in the global initiative in fighting cancer.
Acknowledgments
I would like to thank my mentor Mr. Martin Gomez, Dr. Daphne Ang, and to my parents, Dr. Serafin Serapio, and Dr. Cherry Serapio who all helped me make this project possible!
References:
Chollet, F. (2016). Building powerful image classification models using very little data. Retrieved from https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html.
Martinson, C. (2018). Transfer Learning and Data Augmentation in Keras. Retrieved from https://craigmartinson.com/post/keras-transfer-learning/.
DATASET:
Kather, J.N., Halama, N., Marx, A. (2016). 100,000 histological images of human colorectal cancer and healthy tissue. [Image dataset]. Retrieved from https://zenodo.org/record/1214456.
How I Built a Colorectal Cancer Prediction Platform through Deep Learning was originally published in Towards AI — Multidisciplinary Science Journal on Medium, where people are continuing the conversation by highlighting and responding to this story.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.