Keeping up with ML Research: A Tool to Navigate the ML Innovation Maze
Author(s): Alessandro Amenta
Originally published on Towards AI.
In the fast-paced world of Machine Learning (ML) research, keeping up with the latest findings is crucial and exciting, but let’s be honest — it’s also a challenge. With a constant stream of advancements and new publications, it’s tough to pinpoint the research that matters to you.
The typical conference website is filled with fascinating new papers, yet their interfaces leave much to be desired — they’re often clunky and make it hard to zero in on the content that’s relevant and interesting to you. This can make the search for new papers time-intensive and a bit frustrating.
Enter ML Conference Paper Explorer: your sidekick in navigating the ML paper maze with ease. It’s all about getting you to the papers you need without the hassle.
Why We Built This
The challenge is real: each ML conference presents a crazy number of papers, usually in the order of thousands, often listed one after the other without a practical way to filter through the noise. Often there is no search at all, it’s literally just a list. In an age where ML is reshaping the future, why is accessing its knowledge still so impractical? Official conference websites, while informative, aren’t exactly user-friendly or conducive to discovery.
So, that’s why we built this: a streamlined platform that not only aggregates and organizes papers from all the major ML conferences but also makes finding the papers you are interested in a lot more straightforward.
What the Project Does and Architecture
Overall here’s what the project does: it aggregates all the accepted papers from the latest ML conferences into one database. We use custom built scrapers to collect the papers and turn them into text embeddings to make them searchable and visualize the data.
- Scraping & Fetching: We’ve developed specialized scrapers and fetchers for each conference, for instance, the Openacess scraper and Arxiv fetcher work together to reel in all the accepted papers from ICCV.
- Data Storage: Important paper details — title, abstract, authors, URL, year, and conference name — are saved in a JSON file in the repo (papers_repo.json), ready for quick keyword searches and filtering.
- Embeddings: Using OpenAI’s text embeddings (ada 002), we transform paper titles and abstracts into embeddings, which we store in a vector DB (Pinecone). This enables semantic or unified search.
- Interactive Visualization: Using t-SNE and Bokeh, we plot all of the embeddings in our vectorDB, so that the user can visually navigate through research clusters.
A Glimpse into the Code
Our scrapers are the backbone of the data collection process. Here’s an insight into their architecture:
class Scraper:
def get_publications(self, url):
raise NotImplementedError("Subclasses must implement this method!")
class OpenAccessScraper(Scraper):
def __init__(self, fetcher, num_papers=None):
self.fetcher = fetcher
self.num_papers = num_papers
logger.info("OpenAccessScraper instance created with fetcher %s and num_papers_to_scrape %s", fetcher, num_papers)
def get_publications(self, url, num_papers=None):
logger.info("Fetching publications from URL: %s", url)
try:
response = requests.get(url)
response.raise_for_status()
except requests.exceptions.RequestException as e:
logger.error("Request failed for URL %s: %s", url, e)
return []
soup = BeautifulSoup(response.content, 'html.parser')
papers = []
arxiv_anchors = [anchor for anchor in soup.find_all('a') if 'arXiv' in anchor.text]
logger.debug("Found %d arXiv anchors", len(arxiv_anchors))
# If num_papers_to_scrape is defined, limit the number of papers
if self.num_papers:
arxiv_anchors = arxiv_anchors[:self.num_papers]
logger.info("Limiting the number of papers to scrape to %d", self.num_papers)
for anchor in arxiv_anchors:
title = anchor.find_previous('dt').text.strip()
link = anchor['href']
arxiv_id = link.split('/')[-1]
abstract, authors = self.fetcher.fetch(arxiv_id)
papers.append({'title': title, 'url': link, 'abstract': abstract, 'authors': authors})
logger.info("Successfully fetched %d papers", len(papers))
return papers
-----------------------------------------------------------------------------
class PublicationFetcher(metaclass=ABCMeta):
'''Abstract base class for publication fetchers.'''
@abstractmethod
def fetch(self, publication_id):
'''Fetches the publication content from the source and returns it.'''
raise NotImplementedError("Subclasses must implement this method!")
class ArxivFetcher(PublicationFetcher):
def fetch(self, arxiv_id):
logger.debug(f"Attempting to fetch publication {arxiv_id} from arXiv")
api_url = f"http://export.arxiv.org/api/query?id_list={arxiv_id}"
headers = {
'User-Agent': 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/58.0.3029.110 Safari/537.3'
}
# Implementing retries with exponential backoff
max_retries = 5
retry_delay = 1 # Start with 1 second delay
for attempt in range(max_retries):
try:
response = requests.get(api_url, headers=headers)
response.raise_for_status() # Check for HTTP request errors
logger.debug("Successfully fetched the data on attempt #%d", attempt + 1)
break # Success, exit retry loop
except requests.exceptions.RequestException as e:
logger.warning("Attempt #%d failed with error: %s. Retrying in %d seconds...", attempt + 1, e, retry_delay)
time.sleep(retry_delay)
retry_delay *= 2 # Exponential backoff
else:
# Failed all retries
logger.error("Failed to fetch publication %s after %d attempts.", arxiv_id, max_retries)
return None, None
soup = BeautifulSoup(response.content, 'xml')
entry = soup.find('entry')
abstract = entry.find('summary').text.strip()
authors = [author.find('name').text for author in entry.find_all('author')]
logger.info("Successfully fetched publication %s from arXiv", arxiv_id)
return abstract, authors
Current Progress and Future Plans
This is just the start — version 0 of our project. We’ve already brought together over 10,000 papers from six key conferences. And this is only the beginning; we’re continuously adding and refining features. There’s plenty of room for improvement, and your insights and patience are invaluable to us during this phase of active development.
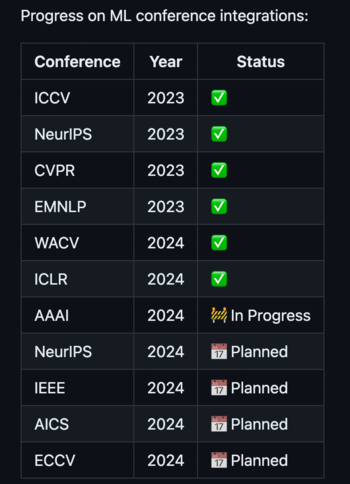
You can give the current version a spin here, or dive into the codebase. We update regularly, so check back often to see the latest updates!
Looking Ahead
Turning papers into embeddings does more than make them easier to find — it helps us spot the big picture. Which research topics are on the rise? What’s the next big thing in ML? Our platform is built to do more than just find papers quickly. It’s about giving you a clearer view of where ML research is heading. Check back with us every month to see new updates and insights we’ve dug up!
Here’s a quick look at app.py, where we bring it all together with a simple Streamlit UI:
import os
import json
import pandas as pd
import streamlit as st
from dotenv import load_dotenv
import streamlit.components.v1 as components
from bokeh.plotting import figure
from bokeh.models import HoverTool, ColumnDataSource
from bokeh.resources import CDN
from bokeh.embed import file_html
from store import EmbeddingStorage
# Load environment variables
load_dotenv()
# Initialize embedding storage with API keys and index name
embedding_storage = EmbeddingStorage(
pinecone_api_key=os.getenv("PINECONE_API_KEY"),
openai_api_key=os.getenv("OPENAI_API_KEY"),
pinecone_index_name="ml-conferences"
)
# Configure the page
st.set_page_config(page_title="ML Conference Papers Explorer U+1F52D", layout="wide")
# Cache and read publications from a JSON file
@st.cache_data
def read_parsed_publications(filepath):
"""Read and parse publication data from a JSON file."""
try:
with open(filepath, 'r') as f:
data = json.load(f)
# Format authors as a comma-separated string
for item in data:
if isinstance(item.get('authors'), list):
item['authors'] = ', '.join(item['authors'])
return data
except FileNotFoundError:
st.error("Publication file not found. Please check the file path.")
return []
# Filter publications based on user query and selections
def filter_publications(publications, query, year, conference):
"""Filter publications by title, authors, year, and conference."""
filtered = []
for pub in publications:
if query.lower() in pub['title'].lower() or query.lower() in pub['authors'].lower():
if year == 'All' or pub['conference_year'] == year:
if conference == 'All' or pub['conference_name'] == conference:
filtered.append(pub)
return filtered
# Perform a unified search combining filters and semantic search
def unified_search(publications, query, year, conference, top_k=5):
"""Combine semantic and filter-based search to find relevant papers."""
filtered = filter_publications(publications, "", year, conference)
if query: # Use semantic search if there's a query
semantic_results = embedding_storage.semantic_search(query, top_k=top_k)
semantic_ids = [result['id'] for result in semantic_results['matches']]
filtered = [pub for pub in filtered if pub['title'] in semantic_ids]
return filtered
# Define file paths and load publications
PUBLICATIONS_FILE = 'papers_repo.json'
existing_papers = read_parsed_publications(PUBLICATIONS_FILE)
# Setup sidebar filters for user selection
st.sidebar.header('Filters U+1F50D')
selected_year = st.sidebar.selectbox('Year', ['All'] + sorted({paper['conference_year'] for paper in existing_papers}, reverse=True))
selected_conference = st.sidebar.selectbox('Conference', ['All'] + sorted({paper['conference_name'] for paper in existing_papers}))
# Main search interface
search_query = st.text_input("Enter keywords, topics, or author names to find relevant papers:", "")
filtered_papers = unified_search(existing_papers, search_query, selected_year, selected_conference, top_k=10)
# Display search results
if filtered_papers:
df = pd.DataFrame(filtered_papers)
st.write(f"Found {len(filtered_papers)} matching papers U+1F50E", df[['title', 'authors', 'url', 'conference_name', 'conference_year']])
else:
st.write("No matching papers found. Try adjusting your search criteria.")
# t-SNE plot visualization
@st.cache_data
def read_tsne_data(filepath):
"""Read t-SNE data from a file."""
with open(filepath, 'r') as f:
return json.load(f)
tsne_data = read_tsne_data('tsne_results.json')
# Assign colors to conferences for visualization
conference_colors = {
'ICLR': 'blue',
'ICCV': 'green',
'NeurIPS': 'red',
'CVPR': 'orange',
'EMNLP': 'purple',
'WACV': 'brown'
}
# Prepare data for plotting
source = ColumnDataSource({
'x': [item['x'] for item in tsne_data],
'y': [item['y'] for item in tsne_data],
'title': [item['id'] for item in tsne_data],
'conference_name': [item['conference_name'] for item in tsne_data],
'color': [conference_colors.get(item['conference_name'], 'grey') for item in tsne_data],
})
# Setup the plot
p = figure(title='ML Conference Papers Visualization', x_axis_label='Dimension 1', y_axis_label='Dimension 2', width=800, tools="pan,wheel_zoom,reset,save")
hover = HoverTool(tooltips=[('Title', '@title'), ('Conference', '@conference_name')])
p.add_tools(hover)
p.circle('x', 'y', size=5, source=source, alpha=0.6, color='color')
# Render the t-SNE plot
html = file_html(p, CDN, "t-SNE Plot")
components.html(html, height=800)
# Add a footer
st.markdown("---")
st.markdown("U+1F680 Made by Alessandro Amenta and Cesar Romero, with Python and lots of U+2764️ for the ML community.")
Here’s what the Streamlit frontend offers: you can apply filters and do semantic searches across all the papers.

And here’s how we display the data: we visualize research paper clusters using t-SNE, making it easy to see how different papers from different conferences are related.
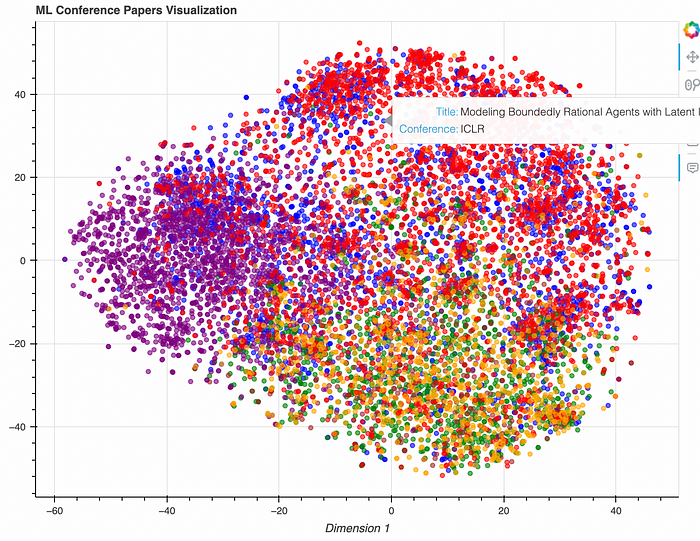
Wrapping up and improvements
We’re nearly ready to open up for contributions and we’d welcome your ideas or feedback even before that. If you’ve got suggestions or improvements, let me know. Help make this tool better for everyone in the ML community. Keep an eye out on the Github repo— we’ll be opening up for contributions in a couple of weeks! U+1F680U+1F50D
Should you find this project useful, consider expressing your appreciation with 50 claps U+1F44F and giving the repo a star U+1F31F— your support means a ton.
Thanks for following along and happy coding! 🙂
Join thousands of data leaders on the AI newsletter. Join over 80,000 subscribers and keep up to date with the latest developments in AI. From research to projects and ideas. If you are building an AI startup, an AI-related product, or a service, we invite you to consider becoming a sponsor.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.