
Introduction To Genomic Data Science
Last Updated on November 12, 2021 by Editorial Team
Author(s): Ömer Özgür
Introduction To Genomic Data Science

Every living organism has its genome at its heart. If the cell is a computer, the genome sequence is the software it executes. If we think of DNA as software run by the cell, we can use our computers to analyze it with similar logic. DNA is not just a storehouse of information.
It is a physical structure that can behave in a complex way. Genomes are incredibly complex machines made up of thousands of parts. Although we know how some genes work today, it is not understood how many genes work together.
- Genetics sees DNA only as information, looking for patterns in data, investigating the relationships between genes and physical appearance.
- On the other hand, Genomics sees the genome as a machine and tries to understand how its parts work together.
Genomic Data Science
Genomic Data Science applies methods such as statistics and machine learning found in data science to genomic problems.
Thanks to next-generation sequencing, we can sequence genomes faster, cheaper, and more successfully. For example, the Human Genome Project was carried out for $2.7 billion, and today you can sequence your genome for $1000. For this reason, petabytes of genomic data can be found on the internet.
A living organism is more complex than any machine that humans have ever produced. Solving this complexity is beyond classical algorithms or human understanding. Most genetic diseases and cancers are caused by the interaction of more than one gene and contain genetic variation.
One of the most potent weapons for analyzing incredibly complex and data point data is Deep Learning, as classical methods assume linear relationships between genes, which is not the case.
With Genomic Data Science, personalized drug studies can be accelerated, or if you’re bored, you can create a 3D version of their faces from their genomes to detect people who have thrown cigarettes or gum on the streets.
Life and DNA
From bacteria to whales, life works on similar principles. All living things are linked together in the tree of life and most likely descended from a single common ancestor.
DNA is a long polymer consisting of 4 basics (A, T, G, C) bases. Almost all information about how to build the organism is stored here.
This information itself and how it is processed (epigenetics) changes over time evolves.
DNA can carry information in near-infinite combinations. The code for people who were never born or for new creatures that never came into existence is waiting to be discovered in this space.
Central Dogma
If DNA is software, proteins are the most critical hardware. Proteins are tiny machines that do most of the work inside the cell. A molecule called mRNA is needed when converting the information in DNA into proteins. The mRNA goes to the Ribosome, and, according to the information it contains, amino acids are combined, and protein is synthesized. Ribosomes are organic 3D printers.
The Breakdown Of Dogma
Now let’s examine how genomes work.
- Eukaryotic DNA is wrapped around proteins called histones to fit inside the cell, tightly packed parts are unreadable, and methylated regions are difficult to read. The mechanisms that regulate when DNA should be opened are not fully understood.
- There is no unidirectional flow of information from DNA to protein. Proteins can also act as regulators by binding to DNA.
- mRNA carries information about which protein is being synthesized, but it does not know when it should be synthesized. Transcription factors come into play here. Transcription factors bind to specific points in DNA and regulate the expression of the genes located nearby.
- miRNA, siRNA, Riboswitches can also participate in the editing task.
Note: Not all editing mechanisms in the genome are understood, and the process is more complex than described here.
Predicting Transcription Factor Binding
We found that a cell is complex and challenging to work with classical methods so that we will practice deep learning techniques. We will use the HepG2 cell line and JunD Transcription factor as data.
HepG2 is an immortal cell line derived from the liver tissue of a 15-year-old African-American child. JunD Transcription factor encoded by the JUND gene. It can downregulate or activate other genes.
We chose the 22nd Chromosome so that the genomic data could be processed. This Chromosome contains about 50 million base pairs. There are about 3 billion base pairs in humans. The largest Chromosome is the 1st, and the smallest is the 22nd.
Genomic data is stored in FASTA or FASTQ format. Data will look like book rows.
>chr22
NNNNNNNNNNNNNNNNNNNNNNNNNNN
TCCCAAATTGTGGAAGGAATGTACATTTGAC
Here we see the base sequences contained in a single strand of DNA. We can ask what the “N” symbol is doing in DNA. This means that it cannot be decided which base is read in the sequencing process.
Next, we need to convert the genomic information into a mathematical format that we can use. As we can see, our data consists of parts A, T, G, C, and N. We can express these parts with One-hot encoding. For example A:[1,0,0,0] or G:[0, 0,1,0] and N:[0.25,0.25,0.25,0.25] can be represented.
[[1., 0., 0., 0.],
[1., 0., 0., 0.],
[0., 1., 0., 0.]
[0., 0., 1., 0.],
[0., 0., 0., 1.]] == ["AATGC"]
After DNA one-hot is encoded, we can start training. Our training data consists of 101 bases and whether the transcription factor is bound to this sequence. For example, if there are 2000 bases, we will have 20 rows. After this point, the problem turns into supervised learning.
Our x[0] == [[1., 0., 0., 0.],
[1., 0., 0., 0.],
[0., 1., 0., 0.]
[0., 0., 1., 0.],
[0., 0., 0., 1.]]..
our y[0] == 1
We can use many models to find the relationship between the x’s and y’s. For this example we will use CNN.
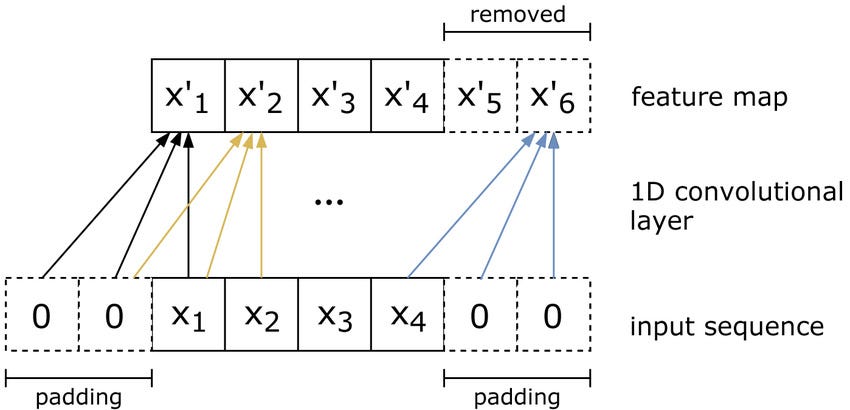
Since DNA is a string, it is one-dimensional. We can use different architectures. One of the most useful architectures for finding patterns in 1D data is Convolutional Neural Networks. They are generally used on images (2D information) and 1D CNNs can be used to process texts.
2D CNNs extract features from the image, learning about edges, vertices, color changes, and more complex patterns. Similarly, we can learn filters that can extract features from text data. Once our model is trained, we can calculate the probability that the JunD protein will bind to DNA fragments 101 bases long.
Generating More Realistic Results
There are many factors in JunD protein binding to DNA: accessibility, methylation, shape, presence of other molecules.
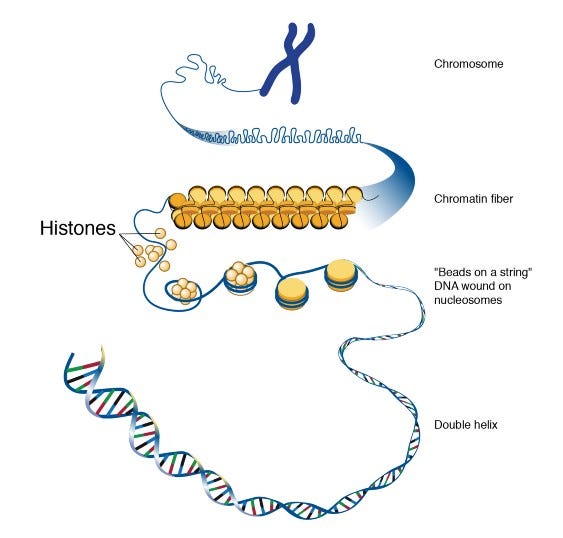
Chromatin accessibility: Defines how accessible the DNA is to molecules from outside. When DNA is tightly wrapped around histones, Transcription Factors or other molecules are inaccessible. Similarly, genes to be silenced can be packaged.
The accessibility of a region is not constant. It changes over time. If we add the chromatin accessibility information to our dataset, we can make more realistic predictions.
Conclusion and Example Code
The complexity of biology is beyond human understanding. This is why we need machine learning to understand and improve ourselves.
When the genome was sequenced, scientists thought they had solved all the secrets, but they had only scratched the surface. Genome editing tools like CRISPR don’t solve our problem either. We cannot edit without predicting the consequences of genetic changes. Unraveling the secret of the genome, if not in an inexplicable way, would provide an opportunity to design new living things to eradicate many diseases.
Data and sample code can be found in my GitHub account.
GitHub – OmerOzgur271/Genomic-Datascience
Data Science was originally published in Towards AI on Medium, where people are continuing the conversation by highlighting and responding to this story.
Published via Towards AI














