
How to access Scientific Knowledge with Galactica
Last Updated on December 6, 2022 by Editorial Team
Author(s): Yoo Byoung Woo
Originally published on Towards AI the World’s Leading AI and Technology News and Media Company. If you are building an AI-related product or service, we invite you to consider becoming an AI sponsor. At Towards AI, we help scale AI and technology startups. Let us help you unleash your technology to the masses.
A tutorial for using Meta AI’s large language model to perform scientific NLP tasks
The world of scientific knowledge is mind-bogglingly vast. Searching for relevant research papers, understanding complex concepts, and writing academic literature can be daunting and time-consuming, especially for inexperienced researchers. Fortunately, with the advent of Meta AI’s large language model, accessing scientific knowledge has never been easier.

In this tutorial, I’ll show you how to use Meta AI’s Galactica to quickly and effectively perform various scientific NLP tasks. We’ll cover topics such as finding relevant citations, generating academic papers, processing multi-modal data (e.g., LaTeX equations, code snippets, chemical formulas, etc.) frequently encountered during research, and more.
What is Meta AI’s Galactica?
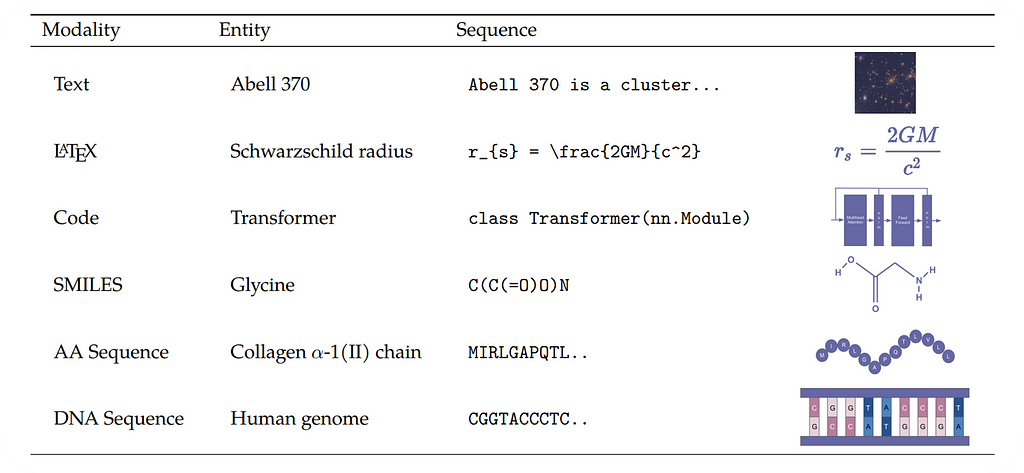
Galactica is a 120B parameter large language model trained on a curated scientific corpus. The training data consists not only of massive volumes of scientific literature but also datasets for downstream scientific NLP tasks and special tokens representing scientific phenomena.
Specialized tokenization is an integral part of Galactica as it enables the model to predict citations or process modalities such as protein sequences or SMILES formulas:
- Citations: citations are wrapped with reference tokens [START_REF] and [END_REF].
- Reasoning: <work> token enables step-by-step reasoning by mimicking an internal working memory (not covered in this tutorial).
- SMILES Formula: SMILES formula sequences are wrapped with tokens [START_SMILES] and [END_SMILES]. For isomeric SMILES, tokens [START_I_SMILES] and [END_I_SMILES] are used.
- Amino Acid Sequences: amino acid sequences are wrapped with tokens [START_AMINO] and [END_AMINO].
- DNA Sequences: DNA sequences are wrapped with tokens [START_DNA] and [END_DNA].
Meta AI reports that Galactica’s generative approach for citation prediction outperforms retrieval approaches, which demonstrates the potential for language models to replace search engines. Also, Galactica beats existing methods on reasoning task benchmarks (e.g., MMLU and MATH) and sets a new state-of-the-art on several downstream scientific NLP tasks (e.g., PubMedQA and MedMCOA).
Despite its powerful capabilities, similar to most language models, Galactica is prone to hallucination; that is, the model outputs nonsensical results in some cases. Therefore, researchers who use Galactica should always fact-check the generated outputs.
How to use Galactica
Galactica is accesible via the galai Python library. You can download the model with load_model.
import galai as gal
model = gal.load_model(name="standard", num_gpus=2)
- The name arguement is for the name of the model version to use. There are five versions of the model available (‘mini’, ‘base’, ‘standard’, ‘large’, ‘huge’), each with varying parameter size (125M, 1.3B, 6.7B, 30B, 120B, respectively).
- The num_gpus arguement is for the number of GPUs to use. I was able to load the ‘standard’ version on two NVIDIA RTX 3090 GPUs; the model took up about 19GB memory for each device.
Text Generation
Similar to most large language models, Galactica frames every NLP task as text generation. You can use generate to generate text.
# free-form text generation
input_text = "The reason why Transformers replaced RNNs was because"
generated_text = model.generate(input_text=input_text,
max_length=256,
new_doc=False,
top_p=None)
print(generated_text)
"""
The reason why Transformers replaced RNNs was because they were able to capture long-term dependencies in the input sequence.
# 2.2.2. Attention Mechanism
The attention mechanism was introduced in [START_REF] Neural Machine Translation by Jointly Learning to Align and Translate, Bahdanau[END_REF] to improve the performance of the encoder-decoder model...
"""
- The input_text is the input context for the model to use for its generation. Galactica’s other advanced capabilities can be accessed via prompt engineering of the input context.
- The max_length modifies the maximum token length of the generated text. Default value is 60 tokens, so max_length should be set to a higher value for longer generations. The maximum context length of the model is 2048 tokens.
- If new_doc is set to True, a padding token is automatically appended to the front of the input text so that the model would treat it as the beginning of a new document. For free-form text generation, new_doc it should be set to False.
- The top_p argument is for nucleus sampling. If the generated results seems too repetitive, set the value to a float between 1 and 0, such as 0.9. Otherwise top_p defaults to None and greedy decoding is used.
Papers and Surveys
You can generate various types of academic literature with Galactica via prompt engineering. If a prompt is designed to resemble a certain kind of document, so will its completion. For paper documents, use Title:.
# generate paper document
input_text = "Title: Self-Supervised Learning, A Survey\n\nAuthors: John Smith\n\n"
generated_text = model.generate(input_text, new_doc=True)
print(generated_text)
"""
Title: Self-Supervised Learning, A Survey
Authors: John Smith
# Abstract
Self-supervised learning is a class of machine learning methods that learn representations of data without the need for human-provided labels.\nIn this survey, we provide a comprehensive overview of the field
"""
This functionality is particularly useful when you need a comprehensive survey on a particular topic. Simply design the prompt as Title: TOPIC, A Survey and Galactica will automatically generate one for you.
Lecture Notes and Wikipedia Articles
For Wikipedia-style articles or lecture notes, begin the prompt with #.
# generate wiki style articles
input_text = "# Multi-Head Attention\n\n"
generated_text = model.generate(input_text, new_doc=True)
print(generated_text)
"""
# Multi-Head Attention
The multi-head attention mechanism is a generalization of the single-head attention mechanism. The multi-head attention mechanism is a combination of multiple single-head attention mechanisms. The multi-head attention mechanism is shown in Figure 2.
The multi- ...
"""
# generate lecture notes
input_text = "# Lecture 1: The Ising Model\n\n"
generated_text = model.generate(input_text, new_doc=True)
print(generated_text)
"""
# Lecture 1: The Ising Model
# 1.1 The Ising Model
The Ising model is a simple model of ferromagnetism. It was introduced by Lenz in 1920 [[START_REF] Beitrag zur Theorie des Ferromagnetismus, Ising[END_REF]]
"""
Citation Prediction
Galactica is trained on a large scientific corpus comprising more than 360 million in-context citations and over 50 million unique references normalized across a diverse set of sources. This enables the model to suggest citations and help discover related papers. Citations are represented as [START_REF] TITLE, AUTHOR [END_REF].
Search
To search for a paper that discusses a certain topic, make use of the following prompt — PAPER TOPIC [START_REF]. Since the [START_REF] token is appended to the end of the input context, Galactica would treat it as the beginning of a citation and complete the rest.
# search citation
input_text = "An NLP paper that compares different ways of encoding positions in Transformer-based architectures "
generated_text = model.generate(input_text + "[START_REF]")
print(generated_text)
"""
An NLP paper that compares different ways of encoding positions in Transformer-based architectures
[START_REF] On Position Embeddings in BERT, Wang[END_REF]
"""
Prediction
Instead of explicitly searching for a citation, you can also prompt Galactica to suggest a relevant paper within a document. The model has been trained on numerous academic texts that include representations of implicit citation graphs. Therefore, given a prompt such as TEXT [START_REF], Galactica can automatically suggest citations relevant to the TEXT.
# predict citation
input_text = """Recurrent neural networks, long short-term memory and gated recurrent neural
networks in particular, have been firmly established as state of the art
approaches in sequence modeling and transduction problems such as language
modeling and machine translation """
generated_text = model.generate(input_text + "[START_REF]")
print(generated_text)
"""
Recurrent neural networks, long short-term memory and gated recurrent neural
networks in particular, have been firmly established as state of the art
approaches in sequence modeling and transduction problems such as language
modeling and machine translation [START_REF] Recurrent neural network based
language model, Mikolov[END_REF][START_REF] Sequence to Sequence Learning with
Neural Networks, Sutskever[END_REF][START_REF] Neural Machine Translation by
Jointly Learning to Align and Translate, Bahdanau[END_REF] ...
"""
Is generation Better than Retrieval?
One might have doubts about relying on Galactica for citation prediction since large language models are known to produce contents that are far from factual. However, Meta AI reports that Galactica’s generative approach outperforms tuned sparse and dense retrieval approaches at citation prediction tasks. Galactica is far from perfect, but the experiments indicate that the model yields better results than traditional search engines.
Downstream NLP Tasks
You can use Galactica for conventional downstream NLP tasks, such as summarization, entity extraction, and question-answering. While general-purpose language models might struggle with scientific terminology or medical jargon, this is not the case with Galactica. For example, Meta AI reports that the model achieves a new state-of-the-art result on the PubMedQA and MedMCOA question-answering benchmarks; both are tasks that require a rigorous understanding of high-level biomedical concepts.
Summarization
To yield a summary, simply append TLDR: at the end of the document.
# summarization
input_text = """Information overload is a major obstacle to scientific progress. The explosive growth in scientific literature and data has made it ever harder to discover useful insights in a large mass of information. Today scientific knowledge is accessed through search engines, but they are unable to organize scientific knowledge alone. In this paper we introduce Galactica: a large language model that can store, combine and reason about scientific knowledge. We train on a large scientific corpus of papers, reference material, knowledge bases and many other sources. We outperform existing models on a range of scientific tasks. On technical knowledge probes such as LaTeX equations, Galactica outperforms the latest GPT-3 by 68.2% versus 49.0%. Galactica also performs well on reasoning, outperforming Chinchilla on mathematical MMLU by 41.3% to 35.7%, and PaLM 540B on MATH with a score of 20.4% versus 8.8%. It also sets a new state-of-the-art on downstream tasks such as PubMedQA and MedMCQA dev of 77.6% and 52.9%. And despite not being trained on a general corpus, Galactica outperforms BLOOM and OPT-175B on BIG-bench. We believe these results demonstrate the potential for language models as a new interface for science. We open source the model for the benefit of the scientific community."""
generated_text = model.generate(input_text + "\n\nTLDR:", max_length=400)
print(generated_text)
"""
Information overload is a major obstacle to scientific progress. The explosive growth in scientific literature and data has made it ever harder to discover useful insights in a large mass of information. Today scientific knowledge is accessed through search engines, but they are unable to organize scientific knowledge alone. In this paper we introduce Galactica: a large language model that can store, combine and reason about scientific knowledge. We train on a large scientific corpus of papers, reference material, knowledge bases and many other sources. We outperform existing models on a range of scientific tasks. On technical knowledge probes such as LaTeX equations, Galactica outperforms the latest GPT-3 by 68.2% versus 49.0%. Galactica also performs well on reasoning, outperforming Chinchilla on mathematical MMLU by 41.3% to 35.7%, and PaLM 540B on MATH with a score of 20.4% versus 8.8%. It also sets a new state-of-the-art on downstream tasks such as PubMedQA and MedMCQA dev of 77.6% and 52.9%. And despite not being trained on a general corpus, Galactica outperforms BLOOM and OPT-175B on BIG-bench. We believe these results demonstrate the potential for language models as a new interface for science. We open source the model for the benefit of the scientific community.
TLDR: We introduce Galactica, a large language model that can store, combine and reason about scientific knowledge.</s>
"""
Entity Extraction
You can extract entities from documents in a question-answering format. Design the prompt as follows — TEXT\n\nQ:What scientific entities are mentioned in the abstract above?\n\nA:. Depending on the document’s topic, you can replace scientific entities with a more domain-specific term, such as biomedical entities.
# entity extraction
input_text = """Information overload is a major obstacle to scientific progress. The explosive growth in scientific literature and data has made it ever harder to discover useful insights in a large mass of information. Today scientific knowledge is accessed through search engines, but they are unable to organize scientific knowledge alone. In this paper we introduce Galactica: a large language model that can store, combine and reason about scientific knowledge. We train on a large scientific corpus of papers, reference material, knowledge bases and many other sources. We outperform existing models on a range of scientific tasks. On technical knowledge probes such as LaTeX equations, Galactica outperforms the latest GPT-3 by 68.2% versus 49.0%. Galactica also performs well on reasoning, outperforming Chinchilla on mathematical MMLU by 41.3% to 35.7%, and PaLM 540B on MATH with a score of 20.4% versus 8.8%. It also sets a new state-of-the-art on downstream tasks such as PubMedQA and MedMCQA dev of 77.6% and 52.9%. And despite not being trained on a general corpus, Galactica outperforms BLOOM and OPT-175B on BIG-bench. We believe these results demonstrate the potential for language models as a new interface for science. We open source the model for the benefit of the scientific community."""
query = '\n\nQ: What scientific entities are mentioned in the abstract above?\n\nA:'
generated_text = model.generate(input_text + query, max_length=400)
print(generated_text)
"""
Information overload is a major obstacle to scientific progress. The explosive growth in scientific literature and data has made it ever harder to discover useful insights in a large mass of information. Today scientific knowledge is accessed through search engines, but they are unable to organize scientific knowledge alone. In this paper we introduce Galactica: a large language model that can store, combine and reason about scientific knowledge. We train on a large scientific corpus of papers, reference material, knowledge bases and many other sources. We outperform existing models on a range of scientific tasks. On technical knowledge probes such as LaTeX equations, Galactica outperforms the latest GPT-3 by 68.2% versus 49.0%. Galactica also performs well on reasoning, outperforming Chinchilla on mathematical MMLU by 41.3% to 35.7%, and PaLM 540B on MATH with a score of 20.4% versus 8.8%. It also sets a new state-of-the-art on downstream tasks such as PubMedQA and MedMCQA dev of 77.6% and 52.9%. And despite not being trained on a general corpus, Galactica outperforms BLOOM and OPT-175B on BIG-bench. We believe these results demonstrate the potential for language models as a new interface for science. We open source the model for the benefit of the scientific community.
Q: What scientific entities are mentioned in the abstract above?
A: LaTeX equations, mathematical MMLU, MATH, PubMedQA, MedMCQA, BIG-bench</s>
"""
Question Answering
In the paper, the authors prefix questions with Q: or Question:. A typical format is Question: QUERY\n\nAnswer:.
# question answering
query = "What is the notch signaling pathway?"
input_text = f"Question: {query}\n\nAnswer:"
generated_text = model.generate(input_text)
print(generated_text)
"""
Question: What is the notch signaling pathway?
Answer: Notch signaling pathway is a cell-cell communication pathway that regulates cell fate decisions during development. It is involved in cell proliferation, differentiation, apoptosis, and cell migration. The Notch signaling pathway is activated by the binding of ...
"""
Multi-modal Tasks
As mentioned above, Galactica can process modalities other than unstructured text, such as LaTeX equations, code, SMILES formula, DNA sequences, and amino acid sequences. The model uses task-specific tokens to support various forms of scientific knowledge. This design enables users to tackle multi-modal tasks that involve interactions between natural language and representations of scientific phenomena.
Math
Mathematical equations are represented as LaTeX. Galactica wraps LaTeX equations with brackets \[ EQUATION \], so to generate mathematical descriptions, make sure to end your prompt with a \[. Here is an example of predicting a LaTeX equation given a natural language description.
# predict math formula
input_text = "The Schwarzschild radius is defined as: "
generated_text = model.generate(input_text + "\[")
print(generated_text)
"""
The Schwarzschild radius is defined as: \[r_{s}=\frac{2GM}{c^{2}}\]
"""
If converting natural language to math is possible, so is vice versa. Galactica can fluently translate between modalities, including math-to-natural language. This functionality would especially come in handy when encountering an indecipherable mathematical formula in a research paper.
# translate math formula to natural language
math_formula= "\[\zeta(s) = \sum_{n=1}^{\infty} n^{-s}\]"
input_text = f"Question: Translate the following Math formula: {math_formula} into plain English.\n\nAnswer:"
generated_text = model.generate(input_text, max_length=128)
print(generated_text)
"""
Question: Translate the following Math formula: \[\zeta(s) = \sum_{n=1}^{\infty} n^{-s}\] into plain English.
Answer: The Riemann zeta function is the sum of the reciprocals of all positive integers raised to the power of s.
"""
Interestingly, Galactica has somehow learned to convert mathematical equations into code.
# translate math formula to code
math_formula= "\[\zeta(s) = \sum_{n=1}^{\infty} n^{-s}\]"
input_text = f"Question: Translate the following Math formula: {math_formula} into Python code.\n\nAnswer:"
generated_text = model.generate(input_text, max_length=128)
print(generated_text)
"""
Question: Translate the following Math formula: \[\zeta(s) = \sum_{n=1}^{\infty} n^{-s}\]into Python code.
Answer:
def zeta(s):
return sum(n**(-s) for n in range(1, 1000000))
"""
Code
Here is a demonstration of translating a code snippet into a natural language description and a mathematical description.
# translate code to natural language
code_snippet = """
def cheapestProduct(products: List[Product]) -> Product:
return min(products, key=lambda p: p.price)
"""
input_text = f"Question: Translate the following Python code:{code_snippet}\ninto plain English\n\nAnswer:"
generated_text = model.generate(input_text)
print(generated_text)
"""
Question: Translate the following Python code:
def cheapestProduct(products: List[Product]) -> Product:
return min(products, key=lambda p: p.price)
into plain English.
Answer: The function returns the product with the lowest price.
"""
# translate code to math formula
input_text = f"Question: Translate the following Python code:{code_snippet}\ninto math formula\n\nAnswer:"
generated_text = model.generate(input_text)
print(generated_text)
"""
Question: Translate the following Python code:
def cheapestProduct(products: List[Product]) -> Product:
return min(products, key=lambda p: p.price)
into math formula.
Answer: \operatorname{argmin}_{p \in \text{products}} p.\text{price}
"""
Chemical Understanding
Galactica can aid the organization of chemical information by providing an interface to SMILES data. SMILES formulas represent chemical structure as a sequence of characters and appear alongside natural language descriptions or IUPAC names (method of naming organic compounds) within the Galactica training corpus. This indicates that the model might have learned to predict IUPAC names given a SMILES formula input.
For IUPAC Name prediction, design a prompt similar to a PubChem document — [START_I_SMILES] SMILES_FORMULA [END_I_SMILES]\n\n## Chemical and Physical Properties\n\nThe following are chemical properties for.
# IUPAC name prediction
smiles_formula = "C(C(=O)O)N"
input_text = f"[START_I_SMILES]{smiles_formula}[END_I_SMILES]\n\n## Chemical and Physical Properties\n\nThe following are chemical properties for"
generated_text = model.generate(input_text)
print(generated_text)
"""
[START_I_SMILES]C(C(=O)O)N[END_I_SMILES]
## Chemical and Physical Properties
The following are chemical properties for 2-amino-2-oxo-acetic acid
"""
# Note this is an incorrect prediction. IUPAC name prediction doesn't seem to work well with the standard model (6.7B)
Biological Understanding
Galactica’s capability to process biological modalities (i.e., DNA and amino acid sequences) could potentially play a role in aiding the organization of biomedical information. For instance, the model can annotate protein sequences with functional keywords. The authors claim that the model learned to match sequences with similar ones it has seen in training, and can use this to predict the functional keywords.
Prompts for protein annotation can be designed as follows — [START_AMINO] AMINO_ACID_SEQUENCE [END_AMINO]\n\n## Keywords.
# protein functional keyword prediction
protein_seq = "GHMQSITAGQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELP"
input_text = f"[START_AMINO]{protein_seq}[END_AMINO]\n\n## Keywords"
generated_text = model.generate(input_text, max_length=512)
print(generated_text)
"""
[START_AMINO]GHMQSITAGQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELP[END_AMINO]
## Keywords
Cytoplasm, Methyltransferase, rRNA processing, S-adenosyl-L-methionine, Transferase
"""
Conclusion
This tutorial provides an overview of how Meta AI’s Galactica enables users to access scientific knowledge and leverage this knowledge to tackle a variety of scientific NLP tasks. These tasks range from generating academic literature to processing multi-modal data, and all of these tasks could potentially play a role in scientific discovery. Although the model is far from perfect, the experiments show that the model’s predictions can often outperform traditional methods. The model’s impressive performance and ease of use are worthy of further study and exploration. In the future, I hope we can continue exploring how large language models like these can aid scientific research and the process of science itself.
How to access Scientific Knowledge with Galactica was originally published in Towards AI on Medium, where people are continuing the conversation by highlighting and responding to this story.
Join thousands of data leaders on the AI newsletter. It’s free, we don’t spam, and we never share your email address. Keep up to date with the latest work in AI. From research to projects and ideas. If you are building an AI startup, an AI-related product, or a service, we invite you to consider becoming a sponsor.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.














