LLMs Encode Clinical Knowledge :A Review
Last Updated on January 19, 2023 by Editorial Team
Author(s): Ronny Polle
Originally published on Towards AI.
LLMs Encode Clinical Knowledge: A Quick Review
Outline
- Introduction
- Contributions
- Limitations
- Conclusion
- References
Introduction
In the field of medicine, language is an enabler of key interactions for and between clinicians, researchers, and patients. This provides opportunities for leveraging LLMs for modeling properties of textual data in the medical domain.
There is evidence that LLMs can act as implicit knowledge bases. The weights of these networks store information, resulting in information that is pliable and hence can be operated upon in a representation space. This phenomenon equips LLMs with an ability to form associations between stored information to produce meaningful insights. The unfortunate news is that this associative capability can lead to hallucinations as information stored in weights is unreliable. Hence, current AI models for applications to medicine and healthcare lack the ability to address significant gaps in effectively leveraging language as a tool for mediating real world clinical workflows. For instance, LLMs are found to have the potential to mirror misinformation,bias, and stereotypes in the corpus.
With the profound advances made by LLMs, AI systems are undergoing innovative repurposing and helping address limitations posed by predominantly single-task AI systems.
Contribution
Key contributions made in this study can be summarised across the following three axes;
- Dataset benchmark
- Framework for human evaluation
- Modeling
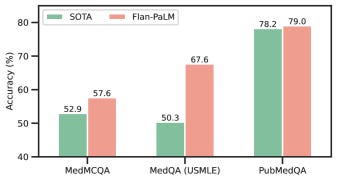
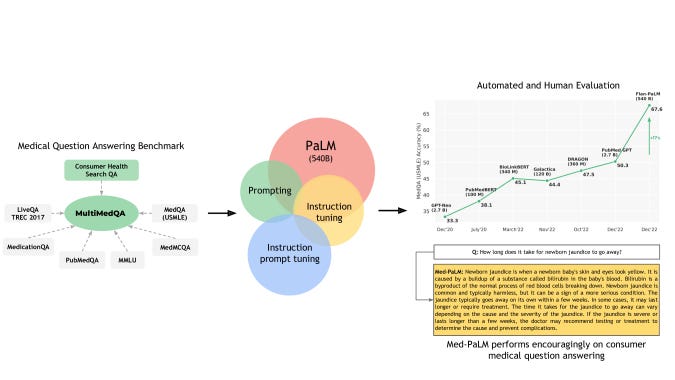
Firstly, this paper introduces a dataset benchmark for medical question answering called MultiMedQA. This benchmark is a collection of six open-question-answering datasets — MedQA [jin2021disease], MedMCQA [pal2022medmcqa], PubMedQA [jin2019pubmedqa], LiveQA [abacha2017overview], MedicationQA [abacha2019bridging], and MMLU clinical topics [hendrycks2020measuring]). MedQA mirrors the US Medical License Exam (USMLE) style of questions. Furthermore, this benchmark is augmented by HealthSearchQA, the seventh dataset comprising curated commonly searched consumer health queries in English.
Secondly, a robust human-centric evaluation framework is proposed to address some of the current limitations with automated metrics for assessing long-form answer generation, such as the bilingual evaluation understanding metric (BLEU). Clinicians and lay users (non-experts) are captured in the evaluation of the models’ generative output. The clinician’s evaluation is run along twelve different evaluation axes whilst lay users are evaluated along two unique axes. These include — how well the models’ output agrees with scientific consensus; the possibility and the likelihood of harm; evidence of comprehension, reasoning, and retrieval ability; the presence of inappropriate, incorrect, or missing content; the possibility of bias in the answer; answer captures user intent and helpfulness of the answer.
Thirdly, this paper highlighted how well LLMs encode clinical knowledge with major architectural modifications. The authors built on the Pathway Language Model (PaLM) and Flan-PaLM family of LLMs. PaLM is a transformer model architecture composed of a decoder-only setup, with key features such as — SwiGLU activation function to substitute for standard activation functions(ReLU, Swish, GeLU), parallel transformer layers, multi-query attention mechanism, rotary position embeddings (ROPE) as a substitute for absolute or relative position embeddings, shared-input output embeddings, zero bias kernels, and layer normalizations, and use of a SentencePiece vocabulary.
Leveraging the PaLM baseline and instruction prompt tuning paradigms, the authors demonstrated the Flan-PaLM variant to gain superior performance across a suite of evaluation tasks over the baseline.

Moreover, given the key limitations of domain adaptation methods and end-to-end finetuning of the model using copious amounts of in-domain data, the authors successfully investigated prompting and prompt-tuning to aid Flan-PaLM in adapting to the medical domain. The instruction prompt tuning technique designed in this study incorporates soft-prompt learned by prompt-tuning as an initial prefix that is shared across multiple medical datasets, followed by a task-specific human-engineered prompt alongside the original question and/or context.

Limitations
Although promising, the dataset benchmark fails to cover multiple languages and excludes a larger variety of medical and scientific domains, hence partially reflecting real-world clinical workflows. Secondly, although Flan-PaLM was able to reach state-of-the-art performance on several medical questions and answering benchmarks, there are important gaps to be bridged in order for it to reach an expert clinician level on many clinically important axes. Important future directions proposed to help address this challenge include — establishing solid grounds for the models’ responses in authoritative medical sources and accounting for the time-varying nature of medical consensus, the ability to effectively quantify and communicate uncertainty to a generic user-in-the-loop, and multilingual support for responses.
Secondly, it is critical to conduct exhaustive work to improve the human evaluation framework. The pilot rating framework is not very exhaustive as it fails to capture important variations across diverse population groups. Also, the pool of clinicians and lay-users assessing the model responses is limited. Lastly, the evaluation framework failed to investigate the impact of variation in the clinician rater’s medical specialty, demographics, and geography.
Moreover, fairness and equity are underexplored in this study, especially the lacking understanding of how perturbations to demographic identifiers in prompts influence the model outputs. Also, the safety-critical and complex requirements of the medical domain pose an important question as to how the approach of sampling clinicians to participate in identifying the best-demonstration prompts examples and crafting few-shot prompts impacts the overall behavior of LLMs.
In conclusion, I am fascinated by the performance of LLMs demonstrated in this rigorous study. Not only does it exemplify a successful application and evaluation of LLMs in the medical context, but also, it demonstrates exciting directions for future research and improvements.
Thank you for reading 🙂
References
[1] Large Language Models (LLMs) encode clinical knowledge
[2] Galactica: A Large Language Model for Science
[3] PaLM : Scaling Language Modeling with Pathways
LLMs Encode Clinical Knowledge :A Review was originally published in Towards AI on Medium, where people are continuing the conversation by highlighting and responding to this story.
Join thousands of data leaders on the AI newsletter. Join over 80,000 subscribers and keep up to date with the latest developments in AI. From research to projects and ideas. If you are building an AI startup, an AI-related product, or a service, we invite you to consider becoming a sponsor.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.














