Automated Annotation of Protein Features Using Language Models
Last Updated on July 17, 2023 by Editorial Team
Author(s): LucianoSphere
Originally published on Towards AI.
At UniProt, ProtNLM is already assisting scientists by connecting protein sequences with descriptions of protein traits in the English language

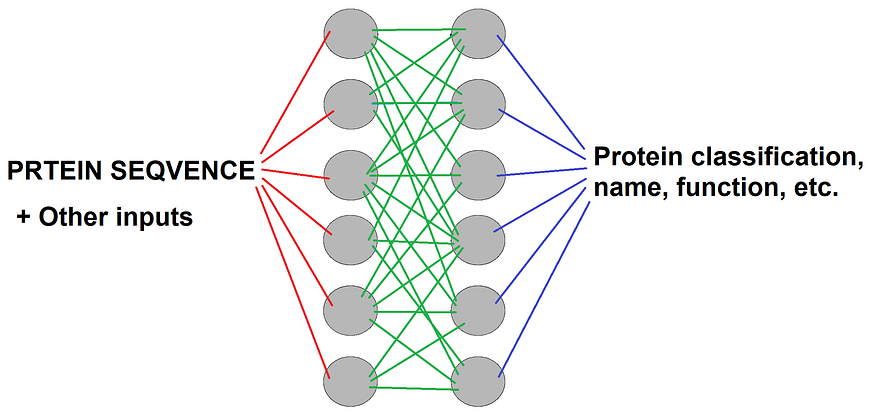
Figure by the author. Note PRTEIN SEQVENCE is not a typo but just reflects the real possible names of the amino acids (O and U don’t exist).
In a nutshell:- ProtNLM is a protein natural language model that is used by UniProt’s Automatic Annotation pipeline for protein annotation.- The model is trained to predict a protein’s name from its amino acid sequence and is based on a sequence-to-sequence model.- ProtNLM can accurately predict descriptions of protein function directly from a protein’s amino acid sequence.- Part of the model was trained to correlate between amino acid sequences obtained from the UniProt database… Read the full blog for free on Medium.
Join thousands of data leaders on the AI newsletter. Join over 80,000 subscribers and keep up to date with the latest developments in AI. From research to projects and ideas. If you are building an AI startup, an AI-related product, or a service, we invite you to consider becoming a sponsor.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.














