From Chaos to Order: Harnessing Data Clustering for Enhanced Decision-Making
Last Updated on August 1, 2023 by Editorial Team
Author(s): Shivamshinde
Originally published on Towards AI.
This article will show the important use cases of data clustering methods, how to use these methods, and also show how one can use these methods as a dimensionality reduction technique.
First of all, let’s discuss the use cases that make these methods so popular.
Customer Segmentation
Online stores use this method for clustering their customers according to their purchasing patterns, day of purchase, age, income, and many more factors. This helps the store to understand its customers better and also helps in making decisions that will ensure high profitability.
To understand this more clearly, let’s take an example. Let’s say after the clustering of the customers according to the day of purchase and their age; we found out that the people with age less than 22 spend quite less during the last days of the month. It is quite likely that most of the crowd with ages less than 22 belong to the students, and since the end of the month is tough financially for most of them, they might be reluctant to visit the store. So, to use this to the store’s advantage, the store might put on end-of-month discount sales, which could pull the student crowd to the store more frequently than before, even at the end of the month.
The store might not have found this solution if they didn’t use clustering on the customers.
You can find one more example of clustering in this Tableau Dashboard.
For data analysis
Sometimes we find some interesting results when we analyze each cluster of the data separately rather than analyzing the whole data together.
As a dimensionality reduction technique
Clustering methods can also be used as dimensionality reduction methods. We will see how to do this at the end of article.
Semi-supervised Learning
We can increase the accuracy of our machine-learning model by first clustering the data and then training a separate model for each cluster.
Sometimes, when we use a semi-supervised learning method for the classification problem, we might get instances with the same label in one of the clusters. To deal with such a situation, we can create a model that returns the same label for every instance that is given as input.
You can find this approach used in one of my projects. You can see the source code for the project on my Github account.
Other use cases
Other than the above-mentioned use cases, clustering is quite helpful in segmenting images, etc.
Now let’s see some of the most famous clustering methods.
KMeans Clustering
KMeans is one of the most famous clustering methods out there. This method will try to find out the blob’s center and then assign each instance to one of the centers.
Let’s see how this method works.
Just start by randomly placing the centroids. Then label each of the clusters. Then assign a label to every instance. The instance will get the label of a cluster that is closest to it. Then we will update the centroids again. After this, we will repeat the process again and again until we find no changes at all to the centroids.
Although this algorithm is guaranteed to converge, it may not converge to the optimal solution. Converging to the right solution depends on the centroid initialization, i.e., the coordinates of the centroid that we use at the start of the algorithm.
One of the solutions to this problem is to run the algorithm multiple times with different random centroid initializations and then keep the best solution. The best solution is found by the performance measure known as inertia. It is basically the mean squared distance between each instance and its closest centroid.
There is a more popular solution to this problem. The new algorithm that implements this solution is known as KMeans++.
It introduced a new initialization step that tends to select centroids that are distant from one another, and this improvement made the kMeans algorithm much less likely to converge to a sub-optimal solution.
There are some more variations of the KMeans algorithm, such as accelerated KMeans or mini — batch KMeans, etc.
We can easily implement this algorithm using the Scikit-Learn library’s built-in classes. But, the real challenge is to find the optimal number of clusters that would separate the data perfectly.
Finding the optimal number of clusters
There are two methods that can be used to find the optimal number of clusters:
- Using the elbow method and a silhouette score
- Using kneed Python library
Using elbow method and silhouette score
The elbow method is basically finding the elbow to the plot between the inertia and the number of clusters. We want an inertia value that is neither too high nor too low. Generally, such value is located at the elbow of the inertia vs number-of-clusters plot.
We can find the silhouette score using the Scikit-learn library. Silhouette's score lies between — 1 and 1.
Silhouette score close to 1 means that the instance is well inside its own cluster and far from other clusters. Silhouette score close to 0 means that it is close to the cluster boundary. Silhouette score close to -1 means that the instance may have been assigned to the wrong cluster.
So we find the optimal number of clusters such that inertia is neither too high nor too low and also silhouette score should also be decent.
Let’s see how to do this.
## necessary imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
## reading the data
"""
For demonstration, we will utilize a straightforward dataset that displays
the amount of tip received by a waiter, based on various factors such
as the day of the week, meal time, total bill, and more.
"""
df = pd.read_csv('https://raw.githubusercontent.com/mwaskom/seaborn-data/master/tips.csv')
df.head()

## plotting the total bill vs tip
plt.figure(figsize=(12,8))
sns.set_style('darkgrid')
plt.plot(df['total_bill'], df['tip'], 'bo')
plt.xlabel('Total Bill')
plt.ylabel('Tip')
plt.title('Total Bill VS Tip')

## clustering the dataframe using the total_bill, tip and size column
## finding the optimal number of clusters using the elbow method
"""
In order to plot the relationship between inertia and number of clusters,
we will train several clustering models with different numbers of clusters
each time. For each of these models, we will record the inertia.
Finally, we will use all the recorded inertia to create a graph.
"""
from sklearn.cluster import KMeans
inertia_list = [KMeans(n_clusters=i).fit(df[['total_bill','tip']]).inertia_ for i in range(2,10)]
plt.figure(figsize=(12,8))
sns.set_style('darkgrid')
plt.plot(list(range(2,10)),inertia_list, 'rx', ls='solid')
plt.xlabel('Number of Clusters')
plt.ylabel('Inertia')
plt.title('Finding the optimal number of clusters using the elbow method')

According to the plot, the optimal number of clusters should be 4 or 5. The inertia value at these two values is neither too high nor too low.
## finding the optimal number of clusters using the silhouette score
from sklearn.metrics import silhouette_score
"""
We will use a similar process to create a plot of silhouette scores vs
number of clusters, as we did while creating the inertia vs number
of clusters plot.
"""
sil_score_list = []
for i in range(2,10):
kmeans = KMeans(n_clusters=i)
kmeans.fit(df[['total_bill','tip']])
sil_score_list.append(silhouette_score(df[['total_bill','tip']],kmeans.labels_))
plt.figure(figsize=(12,8))
sns.set_style('darkgrid')
plt.plot(list(range(2,10)),sil_score_list, 'mx', ls='solid')
plt.xlabel('Number of Clusters')
plt.ylabel('Silhouette Score')
plt.title('Finding the optimal number of clusters using the silhouette score')

According to the two plots above, at the number of clusters equal to 4, we will get a decent enough silhouette score as well as a good inertia value. So, we can use 4 clusters to find out the good performance for the clustering.
Using kneed Python library
## finding the optimal number of clusters for the kmeans model using the kneed library
from kneed import KneeLocator
kn = KneeLocator(range(2,10), inertia_list, curve='convex',direction='decreasing')
print(f"The optimal number of clusters for the kmeans model: {kn.knee}")
Kneed library also gave the same value for the number of clusters as that of the first method. Now let’s use 4 clusters to separate the data and then visualize the clusters.
## using the number of cluster equal to 4 for clustering the data
kmeans_final = KMeans(n_clusters=4)
kmeans_final.fit(df[['total_bill','tip']])
pred = kmeans_final.predict(df[['total_bill','tip']])
## making a new column for cluster labels
df['cluster'] = pred
## visualizing clusters
## plotting the data after clustering
sns.set_style('whitegrid')
sns.jointplot(x='total_bill', y='tip', data=df, hue='cluster',palette='coolwarm');

DBSCAN Clustering
This algorithm defines the clusters as continuous regions of high density separated by regions of low density. Due to this, the clustering made by DBSCAN can take any shape, unlike KMeans, which gives convex-shaped clusters.
The most important component of the DBSCAN algorithm is the concept of core samples. Core samples are the instances present in high-density regions. So basically, clusters in the DBSCAN algorithm are the set of core samples that are close to each other and a set of non-core samples that are close to core samples. We can easily implement the DBSCAN algorithm using the Scikit-Learn DBSCAN class. This class has two important parameters, min_samples, and eps, which define what we mean when we say dense.
For each instance, the algorithm counts how many instances are located within a small distance eps from it. This region is called the instance’s eps — neighborhood.
If an instance has at least min_samples instances in its eps-neighborhood (including itself), then it is considered a core instance. All instances in the neighborhood of the core instance belong to the same cluster. This neighborhood may include other core instances and hence a long sequence of neighboring core instances from a single cluster.
Let’s see how to perform clustering using Scikit-Learn class using the same data that we used for KMeans clustering.
from sklearn.cluster import DBSCAN
## making the clusters
dbscan_pred = DBSCAN(eps=2, min_samples=5).fit_predict(df[['total_bill','tip']])
df['DBSCAN_Pred'] = dbscan_pred
## plotting the data after clustering
sns.set_style('whitegrid')
sns.jointplot(x='total_bill', y='tip', data=df, hue='DBSCAN_Pred',palette='coolwarm');
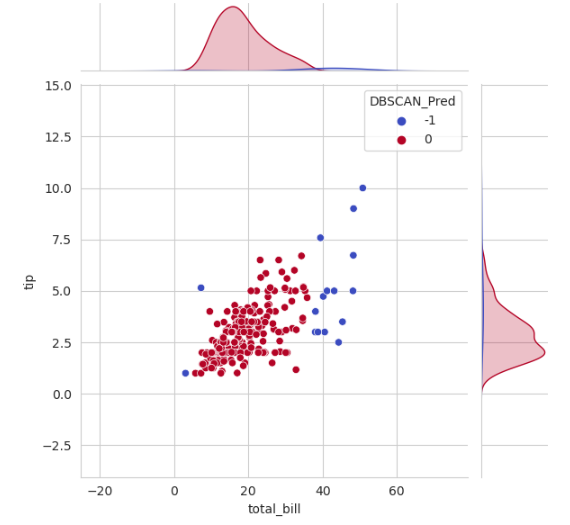
Note that the noisy data samples are given the label -1.
There are many other clustering algorithms such as agglomerative clustering, mean-shift clustering, affinity propagation, spectral clustering, etc.
Now that we have learned how to implement clustering algorithms, let’s see how we can use these methods for the dimensionality reduction problem.
Using clustering as a dimensionality reduction method
We can find out the affinity of each instance with each cluster once the data clustering is completed.
Affinity is the measure of how well each instance fits into different clusters.
Once we have the affinity vector of each instance, we can then replace the original instance with its affinity vector. If the affinity vector is k-dimensional, then the new dimensions of the data will be k only.
It doesn’t matter how many dimensions the original data has, after clustering, the data will have dimensions equal to the number of clusters into which the data is divided.
Let us use an iris flower dataset for this demonstration.
## necessary imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import confusion_matrix
## reading the dataset
df1 = pd.read_csv('data-path')
## Removing unnecessary feature
df1.drop('Id',axis=1,inplace=True)
## Dividing the data into dependent and independent features
X = df1.drop('Species',axis=1)
y = df1['Species']
## Dividing the data into train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y,random_state=238, test_size=0.33)
## let's first train the model directly without using any dimensionality reduction technique
## here let's use the random forest classifier model for the training
rfc = RandomForestClassifier(n_estimators=250,n_jobs=-1,max_depth=3)
rfc.fit(X_train, y_train)
rfc_predictions = rfc.predict(X_test)
## checking the confusion matrix of the predictions
from sklearn.metrics import confusion_matrix
matrix = confusion_matrix(y_test, rfc_predictions)
print("Confusion Matrix for the model created before applying any kind of dimensionality reduction method:\n")
confusion_matrix(y_test,rfc_predictions, labels = df1['Species'].unique())

## necessary imports
from sklearn.cluster import KMeans
## let's create 3 clusters
k = 3
kmeans = KMeans(n_clusters=k)
kmeans.fit(X_train)
X_train_new = kmeans.transform(X_train)
## now using these values as a new training set
rfc_new = RandomForestClassifier(max_depth=3,n_jobs=-1,n_estimators=250)
rfc_new.fit(X_train_new,y_train)
rfc_new_predictions = rfc_new.predict(kmeans.transform(X_test))
## checking the confusion matrix of the predictions after replacing feature vectors for instances with the affinity vector
print("Confusion Matrix for the model created after replacing feature vectors for instances with the affinity vector:\n")
confusion_matrix(y_test,rfc_new_predictions, labels = df1['Species'].unique())

Here we can see that there are 2 more inaccurate predictions than before. This is due to the fact that the dimensionality reduction method losses some information. However, having only 2 inaccurate predictions still indicates a high level of accuracy.
I hope you like the article. If you have any thoughts on the article then please let me know. Any constructive feedback is highly appreciated.
Connect with me on LinkedIn.
Have a great day!
Join thousands of data leaders on the AI newsletter. Join over 80,000 subscribers and keep up to date with the latest developments in AI. From research to projects and ideas. If you are building an AI startup, an AI-related product, or a service, we invite you to consider becoming a sponsor.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.