New AI Method for Protein Structure Prediction Handles All Kinds of Biologically Relevant Molecules
Last Updated on November 5, 2023 by Editorial Team
Author(s): LucianoSphere
Originally published on Towards AI.
Allowing scientists to predict their joint structures and to create new proteins designed to specifically bind defined molecules
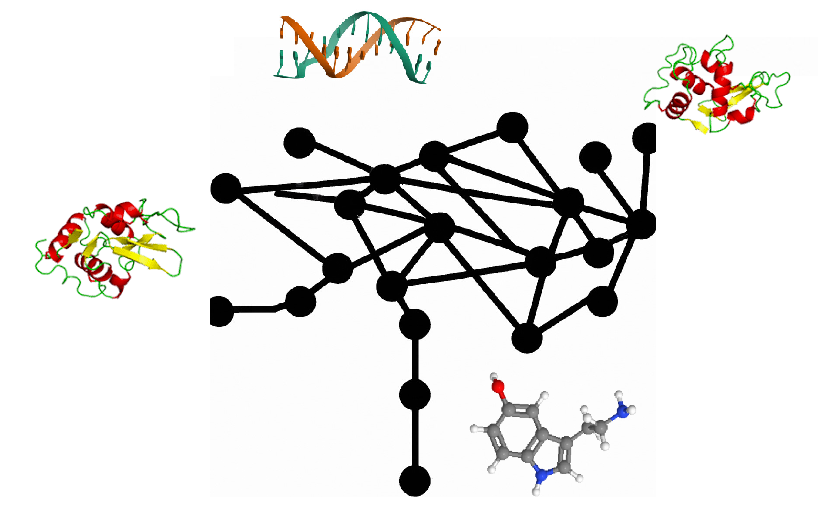
Image composed by the author from Dall-E 2 generations and own-made illustrations.
Predicting the complex three-dimensional structures of proteins with high accuracy is no longer a dream, thanks to deep learning networks like AlphaFold2 and others that followed it. But proteins don’t work alone. They interact with other proteins, with DNA, RNA, and small molecules and ions of all kinds — all crucial to their biological function. These interactions have been a huge challenge to model, but that’s only until now, again, thanks to deep learning.
Presented in a preprint last week, the new software RoseTTAFold All-Atom (I’ll call it just RFAA)… Read the full blog for free on Medium.
Join thousands of data leaders on the AI newsletter. Join over 80,000 subscribers and keep up to date with the latest developments in AI. From research to projects and ideas. If you are building an AI startup, an AI-related product, or a service, we invite you to consider becoming a sponsor.
Published via Towards AI
Take our 90+ lesson From Beginner to Advanced LLM Developer Certification: From choosing a project to deploying a working product this is the most comprehensive and practical LLM course out there!
Towards AI has published Building LLMs for Production—our 470+ page guide to mastering LLMs with practical projects and expert insights!

Discover Your Dream AI Career at Towards AI Jobs
Towards AI has built a jobs board tailored specifically to Machine Learning and Data Science Jobs and Skills. Our software searches for live AI jobs each hour, labels and categorises them and makes them easily searchable. Explore over 40,000 live jobs today with Towards AI Jobs!
Note: Content contains the views of the contributing authors and not Towards AI.















